Dr. Kalpana Surendranath
MSc PhD SFHEA FRSB

Prof. John Murphy
BSc PhD SFHEA FRSB
Application of CRISPR- engineered cellular models in human disease research
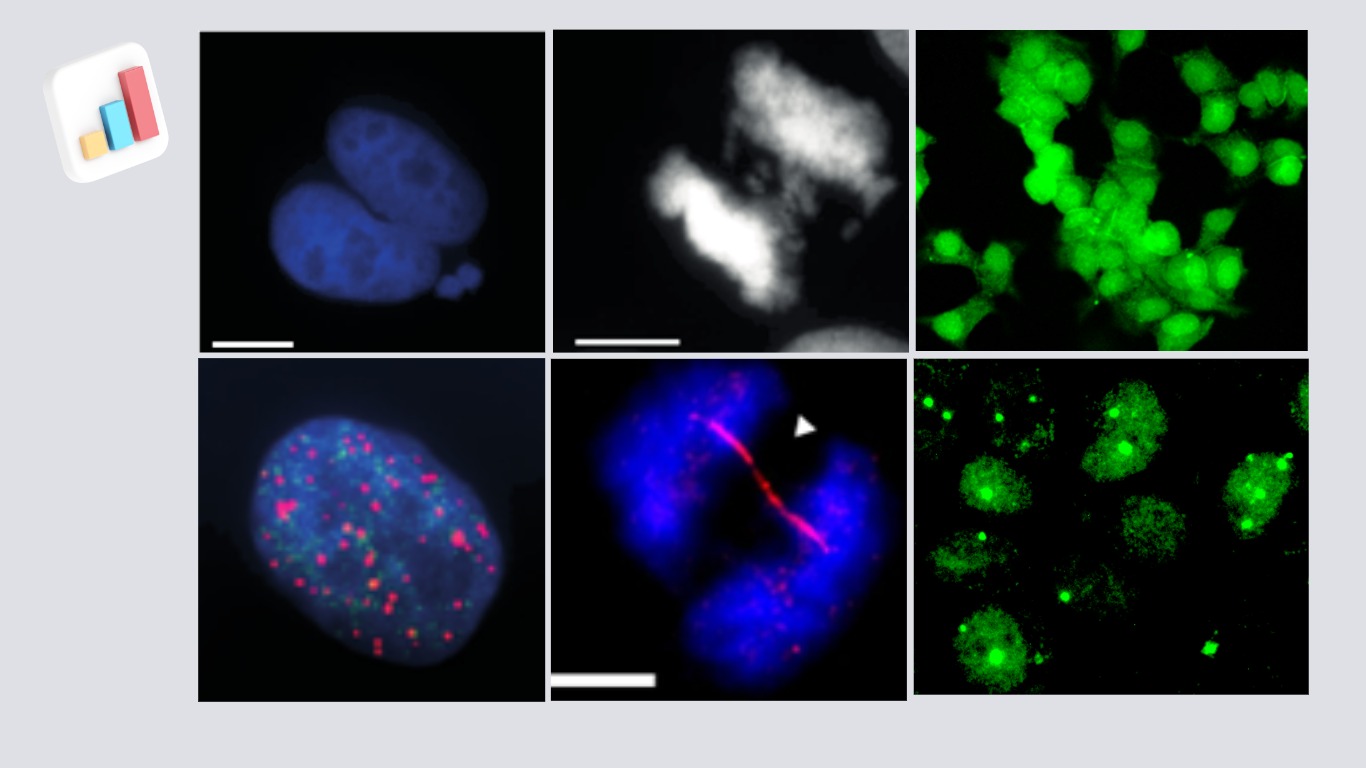
Theme 1 : RNA binding proteins at the crossroads of post-transcriptional gene regulation and genome stability
RNA binding proteins (RBPs) are versatile molecules crucial to various aspects of RNA biology, including biogenesis, function, stability, localization, and transport. Eukaryotic cells produce a plethora of RBPs, each with its unique protein-protein interaction characteristics and RNA-binding capabilities. RNA-binding protein ZFP36L1 is characterised by the CCCH class of tandem zinc finger domains and recognise conserved Adenylate Uridylate-rich elements (AREs) found in the 3′ untranslated regions (UTRs) of mRNAs. They play a pivotal role in promoting mRNA degradation, ultimately leading to mRNA decay. The loss of ZFP36 family members’ expression can lead to the dysregulation of several mRNA targets, some of which are critical for the control of oncogenes and tumour suppressor genes. Despite rapid technological advancements in recent years, there remains limited understanding of the functional relevance of RBPs that are found to be differentially expressed in tumours. While there has been remarkable progress in uncovering RNA-binding motifs and the modes of RBP-RNA interactions, many questions persist, including the structural details of these proteins, their subcellular localization, links to genome instability, their precise arrangements in complex RNA assemblies within different cellular compartments, and their connections to various diseases
Theme 2 : Assessment of DNA Damage Response Vulnerabilities Using CRISPR-Engineered Cellular Models
The health and physiological well-being of an organism hinge on the constant interplay between mechanisms that induce DNA damage and those responsible for its repair. Despite its seemingly protected location, the genetic material, DNA, faces daily threats to its integrity through sources both internal and external to the cell. These threats manifest in diverse ways, encompassing structural modifications and functional alterations of the key components involved in DNA damage response (DDR). Notably, the majority of cancers exhibit heightened reliance on specific DDR mechanisms, often resulting from the loss of one or more DDR functions during oncogenesis. Empirical investigations have underscored an increased rate of mutations in DDR genes within tumour cells, establishing a clear link between replication stress and tumour progression. By pinpointing these dependencies, we can leverage precision medicine strategies and targeted DDR inhibitors to maximise DNA damage, selectively eradicating cancer cells.
Theme 3 : Impact of environmental agents on cellular processes and survival mechanisms
Bisphenols and per- and polyfluoroalkyl substances (PFAS) are prominent environmental pollutants and endocrine-disrupting chemicals with widespread detection in human samples and natural ecosystems. Bisphenols, commonly found in food packaging, toys, water pipes, and medical tubing, are detected in 90% of urine samples globally and represent a significant environmental health concern due to their potential toxicity. Prolonged exposure to bisphenols can inactivate tumour-suppressing genes and activate oncogenes, contributing to breast cancer, one of the most frequently diagnosed cancers among women in developed countries. Similarly, PFAS, which are characterised by their strong C-F bond, high polarity, and low critical micelle concentration (CMC), exhibit high water solubility, bioaccumulation, and biomagnification. The PubChem database identifies over 7 million types of PFAS. Recent studies have mapped PFAS hotspots, revealing elevated concentrations in the waters of China, North America, and Europe. PFAS exposure affects cellular health through multiple mechanisms, including epigenetic alterations such as DNA methylation. However, the molecular mechanisms underlying PFAS exposure in disease remain under-researched.
Theme 4 : CRISPR-based diagnostics
The sudden emergence of the COVID-19 pandemic and the growing demand for reliable diagnostic tools have shifted our focus towards innovations capable of reaching remote parts of the world. CRISPR-based diagnostics, which initially served as basic research tools, have rapidly evolved into efficient, clinically relevant diagnostic platforms. Our goal is to leverage these advancements to develop an improved workflow for creating portable, sensitive, and rapid nucleic acid detection platforms. These platforms will support critical applications, including monitoring disease epidemiology, facilitating diagnostics, and streamlining laboratory tasks that require nucleic acid detection. Aligning with the primary interests of the lab, improvements in this field will be harnessed to enable the monitoring of genetic markers indicative of cancers, further expanding their impact on global healthcare.
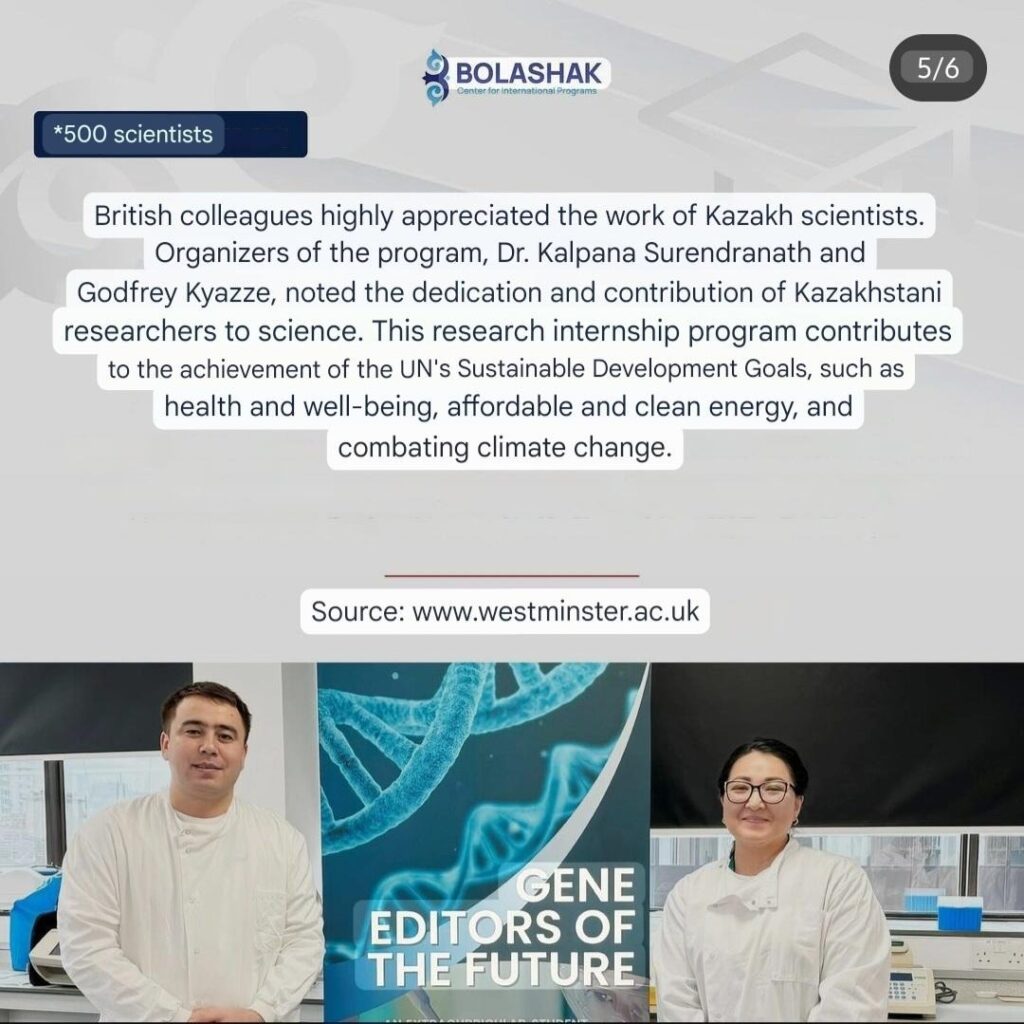
Theme 5 : Gene Editors of the Future: A unique model for research-engaged education at the University of Westminster
Gene editing technologies enable precise and desirable modifications in the genome of living cells and have transformed research and development in biomedical sciences and biotechnology. The Gene Editors of the Future program, hosted by the School of Life Sciences at the University of Westminster is a research-led initiative contributing to advancing academia by fostering innovation and skill development among young researchers. This program unites student researchers passionate about gene editing, providing them with the theoretical foundation and practical expertise necessary to tackle molecular and cellular challenges using cutting-edge CRISPR/Cas9 technology. By offering authentic learning experiences, the program also equips participants with critical skills that prepare them for leadership roles in scientific discovery and application. Since its inception in 2020, the program has engaged over 700 participants including undergraduates, postgraduates, doctoral researchers, postdoctoral fellows, and technicians through co-curricular and extracurricular activities. As a free extracurricular opportunity, it exemplifies the University of Westminster’s commitment to supporting academic growth and fostering collaboration across disciplines. We welcome collaborations to join the Gene Editing Network of Excellence, fostering the exchange of best practices and providing an efficient platform for student-led innovations.

Mr. Khalid Akram
(Phd Student – 3rd Year)
Genomic alterations and functional impacts of CRISPR-Cas9-mediated targeting of ZFP36L1 in breast cancer models.

Mrs. Nahida Bashir
(PhD Student Part-time Final Year)
ZFP36 family of RNA binding proteins: Functional characterization in pathogenesis and progression of cancer

Dr. Nazym Tleumbetov
(Bolashaq international scholar, Westminster Biotechnology Internship programme 2024-2025)

Dr. Aigul Dinmukhamedova
(Bolashaq international scholar, Westminster Biotechnology Internship programme 2024-2025)

Dr. Serzhan Mombekov
(Bolashaq international scholar, Westminster Biotechnology Internship programme 2023-2024)

Dr. Assem Kalykova
(Bolashaq international scholar, Westminster Biotechnology Internship programme 2023-2024)
-
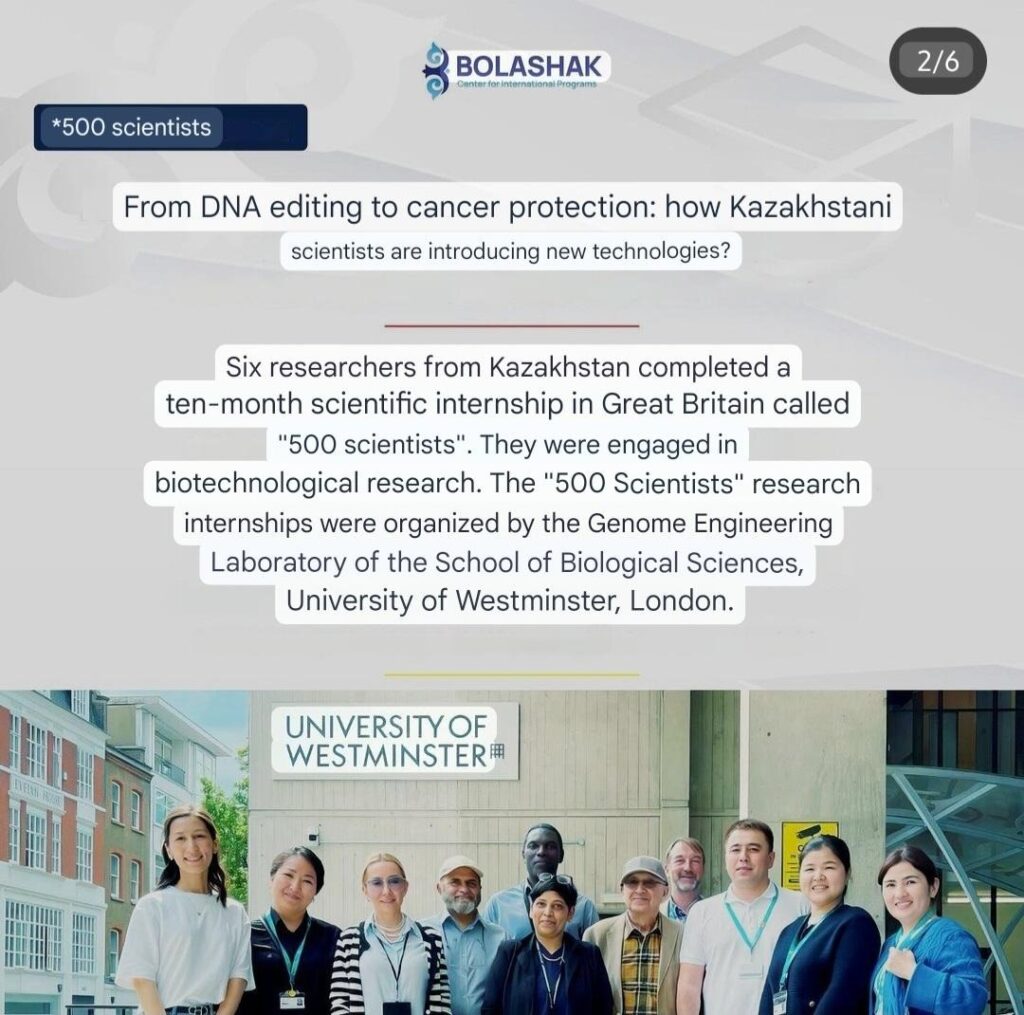
Postdoctoral Bolashaq researchers from Kazakhstan complete Westminster Biotechnology Research Internship Programme

The research fellows, Dr Aknur Turgumbayeva, Dr Assem Kalykova, Dr Elmira Kartbayeva, Dr Gulnaz Seitimova, Dr Gulzira Vassilina and Dr Serzhan Mombekov, […]Read More… from Postdoctoral Bolashaq researchers from Kazakhstan complete Westminster Biotechnology Research Internship Programme
-
Westminster welcomes Bolashak Research Fellows from Kazakhstan

The Bolashak Scholarship Program is a prestigious scholarship initiative in Kazakhstan. Established in 1993, it was designed to support the education of […]Read More… from Westminster welcomes Bolashak Research Fellows from Kazakhstan
Bolashaq 500 scientists international exchange program
Coming Soon…
Coming Soon…

Pragyan Acharya
(Additional Professor, Department of Biochemistry AIIMS international fellowship for faculty)

Dr Nadeen Shaikh Solaiman
(Alumni)

Dr Ahmed Sidali
(Alumni)

Dr Varsha Teotia
(Alumni)
Development activities led by Ahmed and Nadeen




Saini, R., Lakos, N., Beeney, M., Florence, H., Thasneem, S., Nanayakkara, S., and Surendranath, K. (2024) ‘Applications of CRISPR-Cas9 in treatment of HIV and inherited blood disorders: A Systematic Review’. Poster presented at the British Conference of Undergraduate Research (BCUR) 2024, London School of Economics and Political Science, London. Available at: https://info.lse.ac.uk/staff/divisions/Eden-Centre/Assets-EC/Documents/BCUR2024/Posters-BCUR-2024/Reeth-Saini-University-of-Westminster-New-FINAL.pdf
Viessmann, J., Zelve, N., Hijazi, A., Khaparde, A., Rojano, D., Nemade, P., Bojinova, I., Masouleh, A. V. M., Choudhury, S., Murphy, J. J., & Surendranath, K. (n.d.). Gene editors of the future. School of Life Sciences, University of Westminster, London. Retrieved from https://info.lse.ac.uk/staff/divisions/Eden-Centre/Assets-EC/Documents/BCUR2024/Posters-BCUR-2024/Jolina-Viessmann-University-of-Westminster-New-FINAL.pdf
Ruiz Perez, M., Chaurasia, H., Bennacef, I. C., Akhtar, M. M., Mahboob, M., Akhtar, M. M., Benyahia, N., Vasisht, A. K., Zaheer, H., Kiran, K., Valencia Zapata, M., Kalykova, A., Mombekov, S., Murphy, J. J., & Surendranath, K., n.d. Investigating the links between the effect of bisphenol variants in an engineered breast cancer model. Available at: https://info.lse.ac.uk/staff/divisions/Eden-Centre/Assets-EC/Documents/BCUR2024/Posters-BCUR-2024/Magdalena-Ruiz-Perez-University-of-Westminster-New-FINAL.pdf
Surendranath, K., Akram, K., Kanagaraj, R., Ishanzadeh, M.C.S., Khan, S., Pantuzcek, J., Karri, M., Guha, S., Rangan, S.L., Kour, E., Amalanathan, K.R., & Murphy, J.J. (2024). Progress and Prospects in CRISPR Genome Engineering Nucleases. In Translational Research in Biomedical Sciences: Recent Progress and Future Prospects (Chapter 4). SpringerNature. ISBN 978-981-97-1776-7
Teotia, V., Pantuczek, J., Bashir VK, W., Murphy, J., & Surendranath, K. DNA replication stress and the human genome: hurdles, hijacks and cell health. In Cell and Molecular Biology – Annual Volume 2024 IntechOpen. ISBN 978-1-80355-232-3
A deep learning workflow for quantification of Micronuclei in DNA damage studies in cultured cancer cell lines: a proof of principle investigation. Anand Panchbhai, Munuse Ceyda Ishanzadeh, Smarana Pankanti, Ahmed Sidali, Nadeeen Solaiman , Radhakrishnan Kanagaraj, John J Murphy , Kalpana Surendranath* Computer methods in Biomedicine 2023 Apr;232:107447.
RNA-Binding Proteins and Their Emerging Roles in Cancer: Beyond the Tip of the Iceberg. Murphy JJ, Surendranath K, Kanagaraj R. Int J Mol Sci. 2023
Reactive oxygen species induce transcription-dependent replication stress in human cells Andrs et al Nat Commun 2023 Mar 30;14(1):1791.
AU-rich element RNA binding proteins: at the crossroads of post-transcriptional regulation and genome and cellular integrity Sidali A, Teotia V, Solaiman NS, Bashir N, Kanagaraj R, Murphy JJ*, Surendranath K* International Journal of Molecular Sciences 2021 Dec 22;23(1):96.
Reactive oxygen species induce transcription-dependent replication stress in human cells Andrs et al Nature communications (Impact factor 17.6) Under final revision 2023
Sidali, A.; Teotia, V.; Solaiman, N.S.; Bashir, N.; Kanagaraj, R.; Murphy, J.J.; Surendranath, K. AU-Rich Element RNA Binding Proteins: At the Crossroads of Post-Transcriptional Regulation and Genome Integrity. Int. J. Mol. Sci. 2022, 23, 96.
A benzimidazole-based new fluorogenic differential/sequential chemosensor for Cu2+, Zn2+, CN-, P2O74-, DNA, its live-cell imaging and pyrosequencing applications. SellamuthuAnbu, Anup Paul, Kalpana Surendranath, Nadeen Shaikh Solaiman, Armando J.L. Pombeiro, Sensors and Actuators B: Chemical, Volume 337, 2021,129785, ISSN 0925-4005, https://doi.org/10.1016/j.snb.2021.129785 (https://www.sciencedirect.com/science/article/pii/S0925400521003543)
Naphthalimide-phenanthroimidazole incorporated new fluorescent sensor for “turn-on” Cu2+ detection in living cancer cells SellamuthuAnbu, Anup Paul, Kalpana Surendranath, Ahmed Sidali, Armando J.L. Pombeiro, Journal of Inorganic Biochemistry, Volume 220, 2021, 111466, ISSN 0162-0134, https://doi.org/10.1016/j.jinorgbio.2021.111466. (https://www.sciencedirect.com/science/article/pii/S0162013421001136)
Di Marco, S., Hasanova, Z., Kanagaraj, R., et al. (2017). RECQ5 helicase cooperates with MUS81 endonuclease in processing stalled replication forks at common fragile sites during mitosis. Molecular Cell, 66, 658– 671.
D. N. Ofosu DN, Opoku-Okrah C, Nkum B and Murphy J. (2015). The expression of SLAMF7 levels in malignant B cells: A novel therapeutic pathway for patients. Journal of Science and Technology, KNUST April 2015, Vol. 35, 1-10
Zekavati, A., Nasir, A., Alcaraz, A., Aldrovandi, M., Marsh, P., Norton, J.D. and Murphy, J.J (2014).Post-Transcriptional Regulation of BCL2 mRNA by the RNA-Binding Protein ZFP36L1 in Malignant B Cells. PLoS ONE 9(7): e102625. doi: 10.1371/journal.pone.0102625.
Nasir A, Norton JD, Baou M, Zekavati A, Bijlmakers M, Thompson S, Murphy JJ (2012). ZFP36L1 negatively regulates plasmacytoid differentiation of BCL1 cells by targeting BLIMP1 mRNA. PLOS One 7(12): e52187. doi:10.1371/journal.pone.0052187.
BaouM, JewellA, Muthurania A, Wickremasinghe RG, YongKL, CarrR, MarshP and MurphyJJ. (2009). Involvement of Tis11b, an AU Rich Binding protein, in induction of apoptosis by Rituximab in B-CLL cells. Leukemia 23, 986–989.
Bell SE, Sanchez MJ, Spasic-Boskovic O, Santalucia T, Gambardella L, Burton GJ, Murphy JJ, Norton JD, Clark AR, Turner M (2006). The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Developmental Dynamics 235,3144-3155.
Rani A, Stebbing J, Giamas G and MurphyJ. (2019). Endocrine resistance in hormone receptor positive breast cancer- from mechanism to therapy. Front. Endocrinology – Cancer Endocrinology (in Press)
Murphy, J.J. and Rani, A. (2016) STAT5 in Cancer and Immunity Journal of Interferon & Cytokine Research 36 (4) 226-237 1079-9907.
Baou M, Norton JD and Murphy JJ (2011). AU-rich RNA binding proteins in hematopoiesis and leukemogenesis. Blood 118, 5732-5740.
Baou M, Jewell A and Murphy JJ (2009). TIS11 family proteins and their roles in post-transcriptional gene regulation. J. Biomedicine and Biotechnology, Article ID 634520, doi:10.1155/2009/634520
Chakravarthi BV, Das P, Surendranath K, Karande AA, Jayabaskaran C. Production of paclitaxel by Fusarium solani isolated from Taxus celebica. J Biosci. 2008 Jun;33(2):259-67. doi: 10.1007/s12038-008-0043-6. PMID: 18535360.
Narayanan S, Surendranath K, Bora N, Surolia A, Karande AA. Ribosome inactivating proteins and apoptosis. FEBS Lett. 2005 Feb 28;579(6):1324-31. doi: 10.1016/j.febslet.2005.01.038. PMID: 15733836
Surendranath K, Karande AA. A neutralizing antibody to the a chain of abrin inhibits abrin toxicity both in vitro and in vivo. Clin Vaccine Immunol. 2008 May;15(5):737-43. doi: 10.1128/CVI.00254-07. Epub 2008 Mar 19. PMID: 18353919; PMCID: PMC2394836
Bagaria A, Surendranath K, Ramagopal UA, Ramakumar S, Karande AA. Structure-function analysis and insights into the reduced toxicity of Abrus precatorius agglutinin I in relation to abrin. J Biol Chem. 2006 Nov 10;281(45):34465-74. doi: 10.1074/jbc.M601777200. Epub 2006 Jun 13. PMID: 16772301
Book Chapters and Monograph
Kalpana Surendranath (2009) Abrus Toxins: Study of the dangerous lectin duo present in the seeds of Indian licorice Lambert academic publishing (LAP), Germany ISBN-13: 978-3838307923
Surendranath, K., Akram, K., Kanagaraj, R., Ishanzadeh, M.C.S., Khan, S., Pantuzcek, J., Karri, M., Guha, S., Rangan, S.L., Kour, E., Amalanathan, K.R., & Murphy, J.J. (2024). Progress and Prospects in CRISPR Genome Engineering Nucleases. In Translational Research in Biomedical Sciences: Recent Progress and Future Prospects (Chapter 4). SpringerNature. ISBN 978-981-97-1776-7
Teotia, V., Pantuczek, J., Bashir VK, W., Murphy, J., & Surendranath, K. DNA replication stress and the human genome: hurdles, hijacks and cell health. In Cell and Molecular Biology – Annual Volume 2024 IntechOpen. ISBN 978-1-80355-232-3